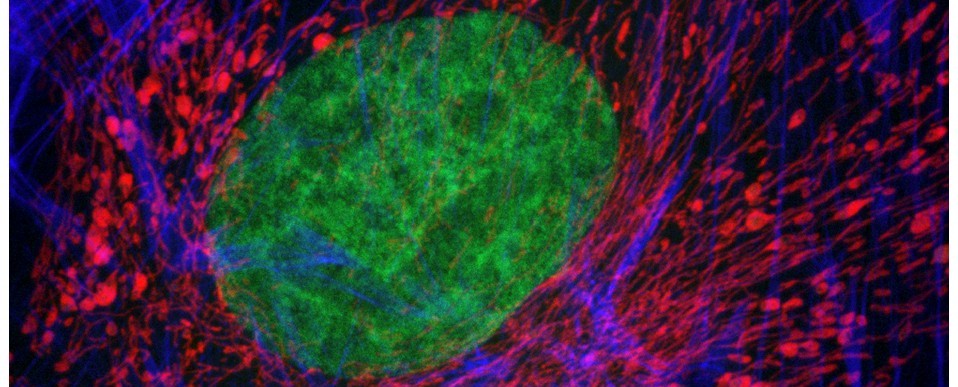
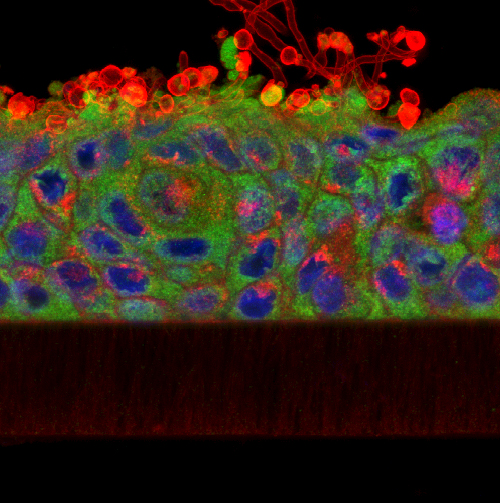
The Zebrafish (Danio rerio) is, in many ways, the perfect model for microscopists. Not only does it share 70% genetic homology with man, but its larvae are born in large, transparent broods all year round and develop extremely quickly (a single cell develops into something resembling a fish within 24 hours!) This means that developmental events can be visualised in vivo in real-time down the microscope. On top of this, their genome has been sequenced and it is easily amenable to molecular manipulation- again, these manipulations can be followed closely under the microscope lens.
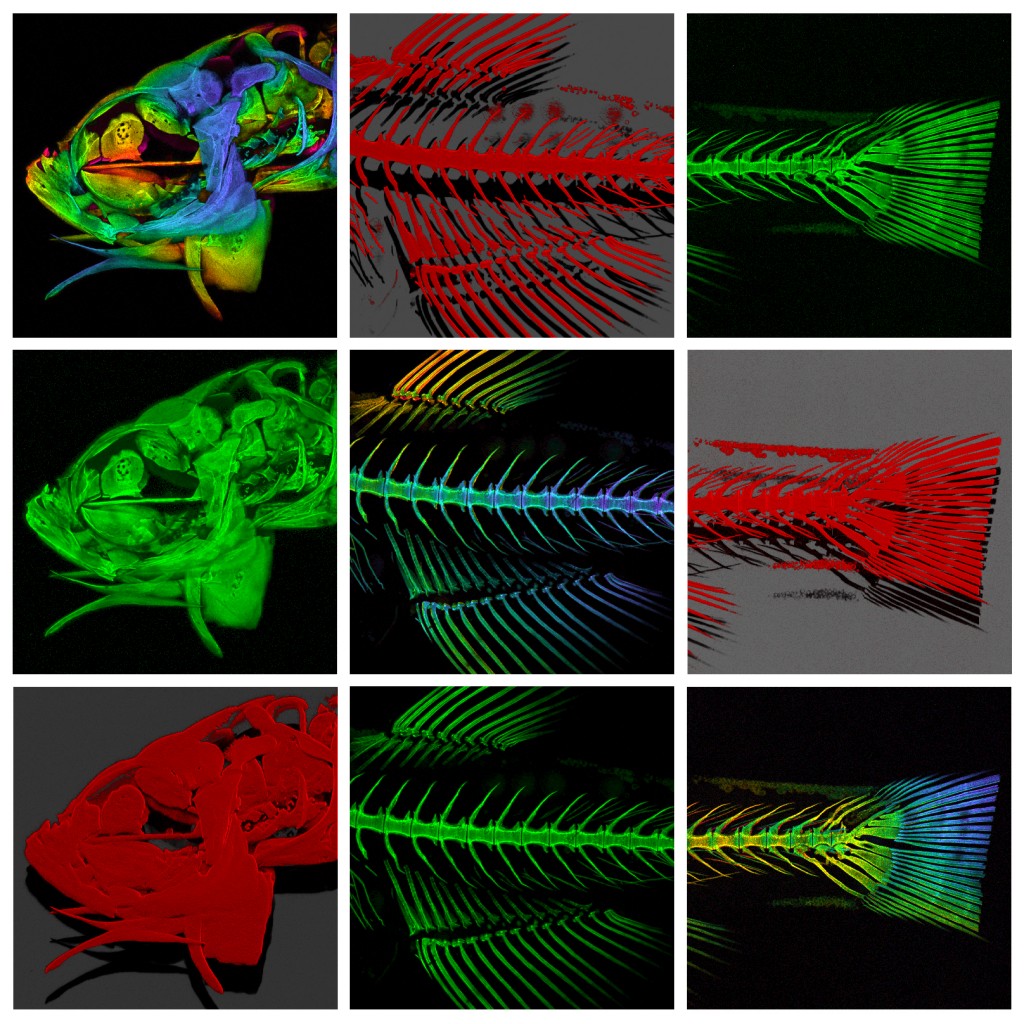
Over the last few years we have been collaborating with Dr Chrissy Hammond at Bristol University, a fish biologist who shares an interest in skeletal development and disease. In our joint studies, we have used a variety of imaging techniques (brightfield, DIC, polarising, epifluorescence, confocal, TEM, radiography and microCT) to investigate skeletal development, growth and ageing in this animal model.
One of the many interesting findings from our studies is that ageing fish undergo degenerative changes to their spine that resemble osteoarthritis (for example, spinal curvature, osteophyte formation, and connective tissue degeneration). This opens up the possibility that they could be used to experimentally model aspects of the human disease. So it’s not just fishy tails!
AJH
Find out more:
- Guardian article: How the diminutive zebrafish is having a big impact on medical research
- Arthritis Research UK: Fishy tales
Further Reading:
- Hayes, et al. (2013) Expression of glycosaminoglycan epitopes during zebrafish skeletogenesis. Developmental Dynamics 242: 778-789
- Hayes et al. (2013) Spinal deformity in aged zebrafish is accompanied by degenerative changes to their vertebrae that resemble osteoarthritis. PLoS ONE 8 : e75787