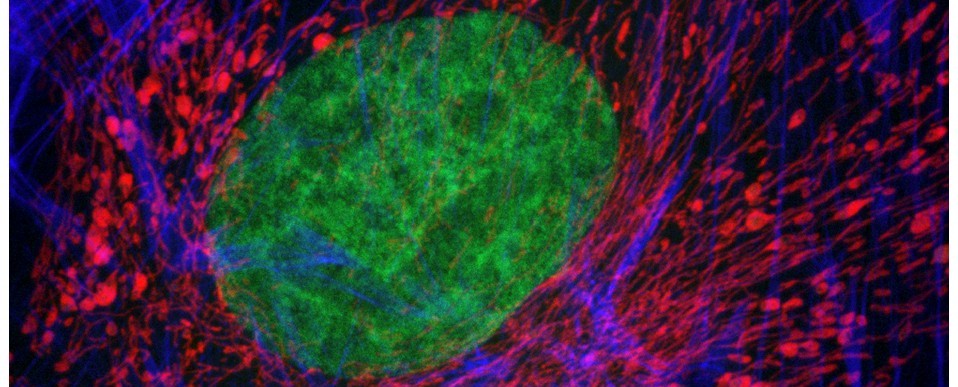
Most of the work of the Bioimaging Hub is concentrated on acquiring images – choosing the right equipment, optimising settings, advising about sample prep, etc. We do, however, have a few systems dedicated to analysing images too. We’ve got a system running Arivis Vision4D which specialises in extremely large datasets, such as Lightsheet data, as well as a system running Bitplane Imaris. We’ve had Imaris for longer and so it’s seen a lot more use. This was recently updated with a Filament Tracer module for analysing neuronal processes. Shortly after this upgrade was added we experienced a severe slowdown in the software. It would take over a minute to go from the ‘Surpass’ view, where images are analysed, to the ‘Arena’ view, where files are organised. The information for the files in Arena is stored in a database and we suspected that the database was at fault.
Imaris hanging when switching from Surpass to Arena. It would do this for about 65 seconds every time a change was made.
A call with Imaris technical support was arranged and the software examined. There were no apparent errors in any of the logs and the database was functioning as it should. The only advice available was to thin down the number of entries in the database – we were told it had nearly 240,000 entries which, even accounting for metadata, seemed vastly excessive for the number of files in Arena.
Complete database dump of nearly 240,000 entries.
I decided to try to trim the database.
My first thought was the Filament Tracer module was generating a large amount of statistics and these were being stored in the database. A couple of users had been experiencing crashes when accessing these statistics so it was possible that slow database responses were bringing the software down. I backed up all datasets which used the Filament Tracer (a process far more laborious than it should be) and deleted them all from Arena. This dropped the database access time from 65 seconds to 60. Not that much of a result.
My next thought was that the co-ordinates for the vertices of surface renders might have been clogging up the database. We’ve generated quite a lot of 3D models of pollen grains and cells so this was potentially a lot of data. I went through the same laborious process of exporting all these datasets and deleted them. Again, little improvement.
I decided I needed to look at the database directly. The underlying database runs on PostgreSQL as part of the BisQue bioimaging package. Using the pgAdmin tool I began to browse the database to see where the data was held and how it was organised.
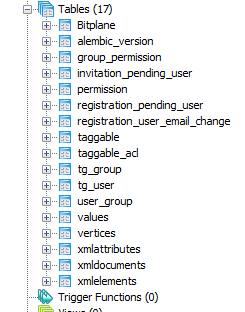
Structure of the underlying database.
I couldn’t find any trace of it so I exported the entire thing as a text file and loaded it into NotePad++. As Imaris technical support had told us, it was enormous – 55MB of text. Scanning the file, eventually I found that practically all the data was held in a database table named ‘taggable’. I’d skipped over this at first as the name was so nondescript.
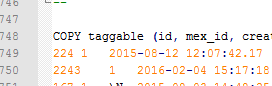
Using Notepad++ to check a database dump and find where the data is stored – the table named ‘taggable’.
Once I knew all the data I needed was in this table I began to examine it. The first thing that jumped out at me was the huge number of entries in the database relating to datasets from our Leica confocal system. This system stores its data as a series of tif images, one per channel, one per z-position for z-stacks. Every single one of these files had its own database entry as a dependency for a ‘resource_parent_id’.
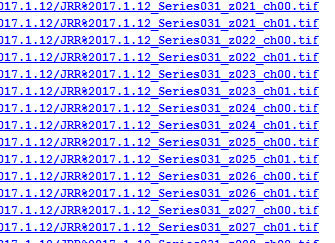
Database entries for Leica datasets. One record per channel, per z-position which becomes a huge number of entries.
A lot of old Leica datasets had been loaded into Arena recently to see if any new information could be extracted from them and this had massively inflated the size of the database. I exported all these datasets as new Imaris .ims files and deleted them from Arena. This reduced the number of database entries from just under 240,000 to just over 16,000. As a result the database access time dropped to about 18 seconds. Much more manageable but still a bit slow.
Looking at the database entries again, I could see that there were still lots of entries relating to Leica datasets. I went back to look at Arena but there was no sign of them. These were orphaned entries relating to non-existent data. As it was impossible to delete them from Arena, I identified all of their resource_parent_id numbers and used pgAdmin to delete them
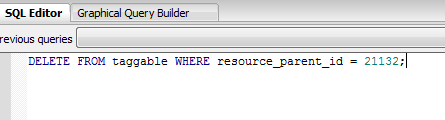
Manually deleting orphaned database entries.
It then occurred to me that the indexes for the database were probably totally out of date so my final task to optimise things was the rebuild all of the indexes in pgAdmin
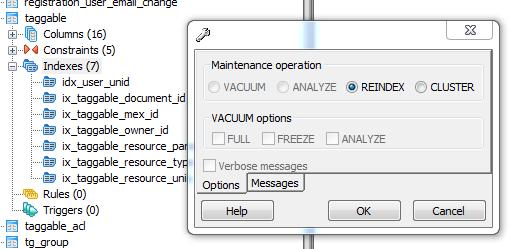
Rebuilding the table indexes.
All of these steps got the database access time down to 3 seconds – quite a bit improvement on the original 65 seconds! Importing some of the exported datasets as Imaris .ims files slowed it back down to about 10 seconds so it’s apparent that the database scales very poorly. Still a lot better than when the Leica datasets were numerous separate files though. It looks to me that the database design favours flexibility over scaling which ends up being not very useful if you want to use it to organise a reasonable amount of imaging data.
So if you’ve got Imaris database lag there’s a few things you can try. The main improvement was to make sure your datasets are represented by single files, either by exporting them as Imaris .ims files or converting them to something like OME-TIFF first.
—
Marc Isaacs, Bioimaging Technician